CRESTはGrimmeグループによって開発された配座探索ツール。半経験的量子力学計算法GFNn-xTBのためのユーティリティーとして開発された。
参考サイト: ComputerChem (2025-05-19) CRESTインストール, Qiita。
CRESTのインストール
コンパイル済みのバイナリをダウンロードして /usr/local/bin にコピーして、sudo chmod +x /usr/local/bin/crest。
xTBのインストール
conda-forgeからインストールしました。
$ conda config --add channels conda-forge $ conda install xtb $ conda activate base
配座探索の実行
9員環ラクタム化合物Indolactam-Vの配座探索を行います。
ILV.xyz
45 C -0.81100 -1.14800 -0.48800 C -0.69700 -2.34500 -1.22600 C -0.95300 -3.59200 -0.66600 C -1.34200 -3.70100 0.66400 C -1.44600 -2.52400 1.41300 C -1.18300 -1.24800 0.89400 C -1.34300 -0.32300 2.00500 C -1.75000 -1.06700 3.09700 N -1.81300 -2.37700 2.72900 N -0.57600 0.04600 -1.16600 C -1.73400 0.85100 -1.51900 C 0.81000 0.55600 -1.30800 C 1.09600 1.12200 -2.72000 C 2.55800 1.57700 -2.84200 C 0.82000 0.08600 -3.81700 C 1.00000 1.61000 -0.18900 O 0.88500 2.82200 -0.34800 N 1.13800 1.01900 1.05800 C 0.38400 1.56600 2.18200 C -1.10500 1.16600 2.05200 C 1.00800 1.11200 3.50900 O 2.33200 1.63400 3.63300 H -0.42000 -2.29200 -2.27700 H -0.86300 -4.48600 -1.27800 H -1.55500 -4.67200 1.10000 H -1.98700 -0.76900 4.11000 H -2.07500 -3.14200 3.33600 H -2.15200 0.50500 -2.47000 H -1.47500 1.90900 -1.63000 H -2.52700 0.78900 -0.76700 H 1.52300 -0.26100 -1.12300 H 0.46200 1.99600 -2.91100 H 3.24800 0.74700 -2.65500 H 2.79300 2.37800 -2.13400 H 2.76100 1.96400 -3.84700 H 1.41300 -0.82200 -3.66200 H 1.07400 0.48900 -4.80400 H -0.23500 -0.19800 -3.84800 H 1.26400 0.01200 1.06600 H 0.45200 2.66000 2.12400 H -1.53600 1.62600 1.15700 H -1.66100 1.59400 2.89800 H 1.08100 0.02200 3.57300 H 0.42500 1.47300 4.36200 H 2.71400 1.63300 2.73500
$ crest ILV.xyz -xnam xtb -gfn2 --gbsa h2o -niceprint -ewin 6 -T 18 > crest.out
-gfn2 はGFN2-xTB法を指定するオプション、-gbsa h2o は水溶媒和モデル、-ewinはエネルギー閾値を6 kcal/mol以内と指定する、-T は計算に使用するCPU(プロセス)の数。
╔════════════════════════════════════════════╗
║ ___ ___ ___ ___ _____ ║
║ / __| _ \ __/ __|_ _| ║
║ | (__| / _|\__ \ | | ║
║ \___|_|_\___|___/ |_| ║
║ ║
║ Conformer-Rotamer Ensemble Sampling Tool ║
║ based on the xTB methods ║
║ ║
╚════════════════════════════════════════════╝
Version 3.0.2, Sun, 25 August 20:02:44, 08/25/2024
commit (af7eb99) compiled by 'usr@fv-az732-492'
Cite work conducted with this code as
• P.Pracht, F.Bohle, S.Grimme, PCCP, 2020, 22, 7169-7192.
• S.Grimme, JCTC, 2019, 15, 2847-2862.
• P.Pracht, S.Grimme, C.Bannwarth, F.Bohle, S.Ehlert,
G.Feldmann, J.Gorges, M.Müller, T.Neudecker, C.Plett,
S.Spicher, P.Steinbach, P.Wesołowski, F.Zeller,
J. Chem. Phys., 2024, 160, 114110.
for works involving QCG cite
• S.Spicher, C.Plett, P.Pracht, A.Hansen, S.Grimme,
JCTC, 2022, 18 (5), 3174-3189.
• C.Plett, S. Grimme,
Angew. Chem. Int. Ed. 2023, 62, e202214477.
for works involving MECP screening cite
• P.Pracht, C.Bannwarth, JCTC, 2022, 18 (10), 6370-6385.
Original code
P.Pracht, S.Grimme, Universität Bonn, MCTC
with help from (alphabetical order):
C.Bannwarth, F.Bohle, S.Ehlert, G.Feldmann, J.Gorges,
S.Grimme, C.Plett, P.Pracht, S.Spicher, P.Steinbach,
P.Wesolowski, F.Zeller
Online documentation is available at
https://crest-lab.github.io/crest-docs/
This program is distributed in the hope that it will be useful,
but WITHOUT ANY WARRANTY; without even the implied warranty of
MERCHANTABILITY or FITNESS FOR A PARTICULAR PURPOSE. See the
GNU Lesser General Public License (LGPL) for more details.
Command line input:
$ crest ILV.xyz -xnam xtb -gfn2 --gbsa h2o -niceprint -ewin 6 -T 14
-xnam :
xtb executable was set to: "xtb"
--gfn2 : Use of GFN2-xTB requested.
--gbsa h2o : implicit solvation
-ewin 6
-T 14 (CPUs/Threads selected)
> Setting up backup calculator ... done.
----------------
Calculation info
----------------
> User-defined calculation level:
: xTB calculation via tblite lib
: GFN2-xTB level
: Molecular charge : 0
: Solvation model : gbsa
: Solvent : h2o
: Fermi temperature : 300.00000
: Accuracy : 1.00000
: max SCC cycles : 500
-----------------------------
Initial Geometry Optimization
-----------------------------
Geometry successfully optimized.
┍━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━┑
│ CREST iMTD-GC SAMPLING │
┕━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━┙
Input structure:
45
C 0.1922437661 -1.4960850248 0.0897334052
C 0.1765463882 -2.6657533739 -0.6543415539
C 1.3295594653 -3.2493639719 -1.1744318231
C 2.5606084738 -2.6770451414 -0.9517564360
C 2.5945403662 -1.4854320276 -0.2377176468
C 1.4331395855 -0.8412836507 0.2758770275
C 1.8964937752 0.3888453894 0.8612729269
C 3.2543375541 0.4085382022 0.7007920192
N 3.6765632245 -0.7114941550 0.0538032076
N -1.0528228940 -1.0836946587 0.6188187230
C -1.1787274445 -0.9476079688 2.0525049206
C -1.9511355590 -0.3331089640 -0.2470551535
C -3.4306980816 -0.6721824436 -0.0296275657
C -4.2845601548 0.1353496855 -1.0045996295
C -3.6688215434 -2.1672177207 -0.2165686547
C -1.6074850447 1.1167745115 0.0813008749
O -2.2363729667 1.7982361301 0.8830666458
N -0.4274856348 1.5248002587 -0.4571280921
C 0.4535273222 2.3853694690 0.3185017027
C 1.1306954393 1.5436404294 1.4254560819
C 1.4667394510 3.0543306425 -0.6111273452
O 0.8755041977 3.9949227158 -1.4755551157
H -0.7763311441 -3.1484394964 -0.8078943880
H 1.2511465285 -4.1694850641 -1.7336596417
H 3.4696272194 -3.1242311862 -1.3228201397
H 3.9477298023 1.1666291388 1.0086506145
H 4.6292832413 -0.9324505017 -0.1806308990
H -1.8417807629 -1.7271886071 2.4455576583
H -1.5953145525 0.0204201959 2.3444377595
H -0.2022700275 -1.0698724762 2.5199514383
H -1.6782855227 -0.5653918746 -1.2818674551
H -3.7010877085 -0.3839094088 0.9904693788
H -4.0132778030 -0.0960479717 -2.0329186108
H -4.1447163659 1.2008555427 -0.8383277713
H -5.3368706205 -0.0977202351 -0.8640771554
H -3.4354617237 -2.4597558056 -1.2385613186
H -4.7101179143 -2.4085161108 -0.0186702232
H -3.0435459024 -2.7419756406 0.4607845134
H 0.0708404132 0.8313891623 -1.0015943253
H -0.1748375933 3.1525879478 0.7826715116
H 0.3487010001 1.2078930708 2.1068466172
H 1.8139012173 2.1844731503 1.9841222489
H 1.9973740977 2.2831782844 -1.1884513374
H 2.1961422036 3.6109013681 -0.0196292060
H 0.1289032220 3.5745789069 -1.9205507845
------------------------------------------------
Generating MTD length from a flexibility measure
------------------------------------------------
Calculating GFN0-xTB WBOs ... done.
Calculating NCI flexibility ... done.
covalent flexibility measure : 0.228
non-covalent flexibility measure : 0.688
flexibility measure : 0.278
t(MTD) / ps : 8.0
Σ(t(MTD)) / ps : 112.0 (14 MTDs)
-----------------------------------
Starting trial MTD to test settings
-----------------------------------
Trial MTD 1 runtime (1.0 ps) ... 0 min, 11.723 sec
Estimated runtime for one MTD (8.0 ps) on a single thread: 1 min 34 sec
Estimated runtime for a batch of 14 MTDs on 14 threads: 1 min 34 sec
******************************************************************************************
** N E W I T E R A T I O N C Y C L E **
******************************************************************************************
------------------------------
Meta-Dynamics Iteration 1
------------------------------
list of applied metadynamics Vbias parameters:
$metadyn 0.13500 1.300
$metadyn 0.06750 1.300
$metadyn 0.03375 1.300
$metadyn 0.13500 0.780
$metadyn 0.06750 0.780
$metadyn 0.03375 0.780
$metadyn 0.13500 0.468
$metadyn 0.06750 0.468
$metadyn 0.03375 0.468
$metadyn 0.13500 0.281
$metadyn 0.06750 0.281
$metadyn 0.03375 0.281
$metadyn 0.04500 0.100
$metadyn 0.22500 0.800
::::::::::::: starting MTD 8 :::::::::::::
| MD simulation time : 8.0 ps |
| target T : 300.0 K |
| timestep dt : 5.0 fs |
| dump interval(trj) : 100.0 fs |
| SHAKE algorithm : true (all bonds) |
| dump interval(Vbias) : 1.00 ps |
| Vbias prefactor (k) : 0.0675 Eh |
| Vbias exponent (α) : 0.4680 bohr⁻² |
::::::::::::: starting MTD 11 :::::::::::::
| MD simulation time : 8.0 ps |
| target T : 300.0 K |
| timestep dt : 5.0 fs |
| dump interval(trj) : 100.0 fs |
| SHAKE algorithm : true (all bonds) |
| dump interval(Vbias) : 1.00 ps |
| Vbias prefactor (k) : 0.0675 Eh |
| Vbias exponent (α) : 0.2808 bohr⁻² |
::::::::::::: starting MTD 9 :::::::::::::
| MD simulation time : 8.0 ps |
| target T : 300.0 K |
| timestep dt : 5.0 fs |
| dump interval(trj) : 100.0 fs |
| SHAKE algorithm : true (all bonds) |
| dump interval(Vbias) : 1.00 ps |
| Vbias prefactor (k) : 0.0338 Eh |
| Vbias exponent (α) : 0.4680 bohr⁻² |
::::::::::::: starting MTD 14 :::::::::::::
| MD simulation time : 8.0 ps |
| target T : 300.0 K |
| timestep dt : 5.0 fs |
| dump interval(trj) : 100.0 fs |
| SHAKE algorithm : true (all bonds) |
| dump interval(Vbias) : 1.00 ps |
| Vbias prefactor (k) : 0.2250 Eh |
| Vbias exponent (α) : 0.8000 bohr⁻² |
::::::::::::: starting MTD 7 :::::::::::::
| MD simulation time : 8.0 ps |
| target T : 300.0 K |
| timestep dt : 5.0 fs |
| dump interval(trj) : 100.0 fs |
| SHAKE algorithm : true (all bonds) |
| dump interval(Vbias) : 1.00 ps |
| Vbias prefactor (k) : 0.1350 Eh |
| Vbias exponent (α) : 0.4680 bohr⁻² |
::::::::::::: starting MTD 13 :::::::::::::
| MD simulation time : 8.0 ps |
| target T : 300.0 K |
| timestep dt : 5.0 fs |
| dump interval(trj) : 100.0 fs |
| SHAKE algorithm : true (all bonds) |
| dump interval(Vbias) : 1.00 ps |
| Vbias prefactor (k) : 0.0450 Eh |
| Vbias exponent (α) : 0.1000 bohr⁻² |
::::::::::::: starting MTD 2 :::::::::::::
| MD simulation time : 8.0 ps |
| target T : 300.0 K |
| timestep dt : 5.0 fs |
| dump interval(trj) : 100.0 fs |
| SHAKE algorithm : true (all bonds) |
| dump interval(Vbias) : 1.00 ps |
| Vbias prefactor (k) : 0.0675 Eh |
| Vbias exponent (α) : 1.3000 bohr⁻² |
::::::::::::: starting MTD 5 :::::::::::::
| MD simulation time : 8.0 ps |
| target T : 300.0 K |
| timestep dt : 5.0 fs |
| dump interval(trj) : 100.0 fs |
| SHAKE algorithm : true (all bonds) |
| dump interval(Vbias) : 1.00 ps |
| Vbias prefactor (k) : 0.0675 Eh |
| Vbias exponent (α) : 0.7800 bohr⁻² |
::::::::::::: starting MTD 6 :::::::::::::
| MD simulation time : 8.0 ps |
| target T : 300.0 K |
| timestep dt : 5.0 fs |
| dump interval(trj) : 100.0 fs |
| SHAKE algorithm : true (all bonds) |
| dump interval(Vbias) : 1.00 ps |
| Vbias prefactor (k) : 0.0338 Eh |
| Vbias exponent (α) : 0.7800 bohr⁻² |
::::::::::::: starting MTD 10 :::::::::::::
| MD simulation time : 8.0 ps |
| target T : 300.0 K |
| timestep dt : 5.0 fs |
| dump interval(trj) : 100.0 fs |
| SHAKE algorithm : true (all bonds) |
| dump interval(Vbias) : 1.00 ps |
| Vbias prefactor (k) : 0.1350 Eh |
| Vbias exponent (α) : 0.2808 bohr⁻² |
::::::::::::: starting MTD 3 :::::::::::::
| MD simulation time : 8.0 ps |
| target T : 300.0 K |
| timestep dt : 5.0 fs |
| dump interval(trj) : 100.0 fs |
| SHAKE algorithm : true (all bonds) |
| dump interval(Vbias) : 1.00 ps |
| Vbias prefactor (k) : 0.0338 Eh |
| Vbias exponent (α) : 1.3000 bohr⁻² |
::::::::::::: starting MTD 12 :::::::::::::
| MD simulation time : 8.0 ps |
| target T : 300.0 K |
| timestep dt : 5.0 fs |
| dump interval(trj) : 100.0 fs |
| SHAKE algorithm : true (all bonds) |
| dump interval(Vbias) : 1.00 ps |
| Vbias prefactor (k) : 0.0338 Eh |
| Vbias exponent (α) : 0.2808 bohr⁻² |
::::::::::::: starting MTD 1 :::::::::::::
| MD simulation time : 8.0 ps |
| target T : 300.0 K |
| timestep dt : 5.0 fs |
| dump interval(trj) : 100.0 fs |
| SHAKE algorithm : true (all bonds) |
| dump interval(Vbias) : 1.00 ps |
| Vbias prefactor (k) : 0.1350 Eh |
| Vbias exponent (α) : 1.3000 bohr⁻² |
::::::::::::: starting MTD 4 :::::::::::::
| MD simulation time : 8.0 ps |
| target T : 300.0 K |
| timestep dt : 5.0 fs |
| dump interval(trj) : 100.0 fs |
| SHAKE algorithm : true (all bonds) |
| dump interval(Vbias) : 1.00 ps |
| Vbias prefactor (k) : 0.1350 Eh |
| Vbias exponent (α) : 0.7800 bohr⁻² |
*MTD 4 completed successfully ... 2 min, 0.580 sec
*MTD 3 completed successfully ... 2 min, 0.981 sec
*MTD 1 completed successfully ... 2 min, 1.238 sec
*MTD 12 completed successfully ... 2 min, 1.282 sec
*MTD 5 completed successfully ... 2 min, 1.477 sec
*MTD 11 completed successfully ... 2 min, 1.821 sec
*MTD 7 completed successfully ... 2 min, 1.977 sec
*MTD 14 completed successfully ... 2 min, 2.063 sec
*MTD 13 completed successfully ... 2 min, 2.065 sec
*MTD 8 completed successfully ... 2 min, 2.211 sec
*MTD 10 completed successfully ... 2 min, 2.409 sec
*MTD 6 completed successfully ... 2 min, 2.656 sec
*MTD 9 completed successfully ... 2 min, 2.692 sec
*MTD 2 completed successfully ... 2 min, 2.859 sec
======================================
| Multilevel Ensemble Optimization |
======================================
Optimizing all 1106 structures from file "crest_dynamics.trj" ...
----------------------
crude pre-optimization
----------------------
Optimization engine: ANCOPT
Hessian update type: BFGS
E/G convergence criteria: 0.500E-03 Eh, 0.100E-01 Eh/a0
maximum optimization steps: 200
|>0.1% |>7.5% |>15.0% |>22.5% |>30.0% |>37.5% |>45.0% |>52.5% |>60.0% |>67.5% |>75.0% |>82.5% |>90.1% |>97.6% |>100.0%
done.
> 1106 of 1106 structures successfully optimized (100.0% success)
> Total runtime for 1106 optimizations:
* wall-time: 0 d, 0 h, 1 min, 35.498 sec
* cpu-time: 0 d, 0 h, 22 min, 6.834 sec
* ratio c/w: 13.894 speedup
> Corresponding to approximately 0.086 sec per processed structure
CREGEN> running RMSDs ... done.
CREGEN> E lowest : -65.09755
729 structures remain within 12.00 kcal/mol window
----------------------------------
optimization with tight thresholds
----------------------------------
Optimization engine: ANCOPT
Hessian update type: BFGS
E/G convergence criteria: 0.100E-05 Eh, 0.800E-03 Eh/a0
maximum optimization steps: 200
|>0.1% |>10.0% |>20.0% |>30.0% |>40.1% |>50.1% |>60.1% |>70.1% |>80.1% |>90.1% |>100.0%
done.
> 729 of 729 structures successfully optimized (100.0% success)
> Total runtime for 729 optimizations:
* wall-time: 0 d, 0 h, 1 min, 26.478 sec
* cpu-time: 0 d, 0 h, 19 min, 54.857 sec
* ratio c/w: 13.817 speedup
> Corresponding to approximately 0.119 sec per processed structure
CREGEN> running RMSDs ... done.
CREGEN> E lowest : -65.09784
122 structures remain within 6.00 kcal/mol window
------------------------------
Meta-Dynamics Iteration 2
------------------------------
list of applied metadynamics Vbias parameters:
$metadyn 0.13500 1.300
$metadyn 0.06750 1.300
$metadyn 0.03375 1.300
$metadyn 0.13500 0.780
$metadyn 0.06750 0.780
$metadyn 0.03375 0.780
$metadyn 0.13500 0.468
$metadyn 0.06750 0.468
$metadyn 0.03375 0.468
$metadyn 0.13500 0.281
$metadyn 0.06750 0.281
$metadyn 0.03375 0.281
$metadyn 0.04500 0.100
$metadyn 0.22500 0.800
::::::::::::: starting MTD 10 :::::::::::::
| MD simulation time : 8.0 ps |
| target T : 300.0 K |
| timestep dt : 5.0 fs |
| dump interval(trj) : 100.0 fs |
| SHAKE algorithm : true (all bonds) |
| dump interval(Vbias) : 1.00 ps |
| Vbias prefactor (k) : 0.1350 Eh |
| Vbias exponent (α) : 0.2808 bohr⁻² |
::::::::::::: starting MTD 6 :::::::::::::
| MD simulation time : 8.0 ps |
| target T : 300.0 K |
| timestep dt : 5.0 fs |
| dump interval(trj) : 100.0 fs |
| SHAKE algorithm : true (all bonds) |
| dump interval(Vbias) : 1.00 ps |
| Vbias prefactor (k) : 0.0338 Eh |
| Vbias exponent (α) : 0.7800 bohr⁻² |
::::::::::::: starting MTD 5 :::::::::::::
| MD simulation time : 8.0 ps |
| target T : 300.0 K |
| timestep dt : 5.0 fs |
| dump interval(trj) : 100.0 fs |
| SHAKE algorithm : true (all bonds) |
| dump interval(Vbias) : 1.00 ps |
| Vbias prefactor (k) : 0.0675 Eh |
| Vbias exponent (α) : 0.7800 bohr⁻² |
::::::::::::: starting MTD 3 :::::::::::::
| MD simulation time : 8.0 ps |
| target T : 300.0 K |
| timestep dt : 5.0 fs |
| dump interval(trj) : 100.0 fs |
| SHAKE algorithm : true (all bonds) |
| dump interval(Vbias) : 1.00 ps |
| Vbias prefactor (k) : 0.0338 Eh |
| Vbias exponent (α) : 1.3000 bohr⁻² |
::::::::::::: starting MTD 2 :::::::::::::
| MD simulation time : 8.0 ps |
| target T : 300.0 K |
| timestep dt : 5.0 fs |
| dump interval(trj) : 100.0 fs |
| SHAKE algorithm : true (all bonds) |
| dump interval(Vbias) : 1.00 ps |
| Vbias prefactor (k) : 0.0675 Eh |
| Vbias exponent (α) : 1.3000 bohr⁻² |
::::::::::::: starting MTD 8 :::::::::::::
| MD simulation time : 8.0 ps |
| target T : 300.0 K |
| timestep dt : 5.0 fs |
| dump interval(trj) : 100.0 fs |
| SHAKE algorithm : true (all bonds) |
| dump interval(Vbias) : 1.00 ps |
| Vbias prefactor (k) : 0.0675 Eh |
| Vbias exponent (α) : 0.4680 bohr⁻² |
::::::::::::: starting MTD 9 :::::::::::::
| MD simulation time : 8.0 ps |
| target T : 300.0 K |
| timestep dt : 5.0 fs |
| dump interval(trj) : 100.0 fs |
| SHAKE algorithm : true (all bonds) |
| dump interval(Vbias) : 1.00 ps |
| Vbias prefactor (k) : 0.0338 Eh |
| Vbias exponent (α) : 0.4680 bohr⁻² |
::::::::::::: starting MTD 1 :::::::::::::
| MD simulation time : 8.0 ps |
| target T : 300.0 K |
| timestep dt : 5.0 fs |
| dump interval(trj) : 100.0 fs |
| SHAKE algorithm : true (all bonds) |
| dump interval(Vbias) : 1.00 ps |
| Vbias prefactor (k) : 0.1350 Eh |
| Vbias exponent (α) : 1.3000 bohr⁻² |
::::::::::::: starting MTD 11 :::::::::::::
| MD simulation time : 8.0 ps |
| target T : 300.0 K |
| timestep dt : 5.0 fs |
| dump interval(trj) : 100.0 fs |
| SHAKE algorithm : true (all bonds) |
| dump interval(Vbias) : 1.00 ps |
| Vbias prefactor (k) : 0.0675 Eh |
| Vbias exponent (α) : 0.2808 bohr⁻² |
::::::::::::: starting MTD 13 :::::::::::::
| MD simulation time : 8.0 ps |
| target T : 300.0 K |
| timestep dt : 5.0 fs |
| dump interval(trj) : 100.0 fs |
| SHAKE algorithm : true (all bonds) |
| dump interval(Vbias) : 1.00 ps |
| Vbias prefactor (k) : 0.0450 Eh |
| Vbias exponent (α) : 0.1000 bohr⁻² |
::::::::::::: starting MTD 4 :::::::::::::
| MD simulation time : 8.0 ps |
| target T : 300.0 K |
| timestep dt : 5.0 fs |
| dump interval(trj) : 100.0 fs |
| SHAKE algorithm : true (all bonds) |
| dump interval(Vbias) : 1.00 ps |
| Vbias prefactor (k) : 0.1350 Eh |
| Vbias exponent (α) : 0.7800 bohr⁻² |
::::::::::::: starting MTD 7 :::::::::::::
| MD simulation time : 8.0 ps |
| target T : 300.0 K |
| timestep dt : 5.0 fs |
| dump interval(trj) : 100.0 fs |
| SHAKE algorithm : true (all bonds) |
| dump interval(Vbias) : 1.00 ps |
| Vbias prefactor (k) : 0.1350 Eh |
| Vbias exponent (α) : 0.4680 bohr⁻² |
::::::::::::: starting MTD 14 :::::::::::::
| MD simulation time : 8.0 ps |
| target T : 300.0 K |
| timestep dt : 5.0 fs |
| dump interval(trj) : 100.0 fs |
| SHAKE algorithm : true (all bonds) |
| dump interval(Vbias) : 1.00 ps |
| Vbias prefactor (k) : 0.2250 Eh |
| Vbias exponent (α) : 0.8000 bohr⁻² |
::::::::::::: starting MTD 12 :::::::::::::
| MD simulation time : 8.0 ps |
| target T : 300.0 K |
| timestep dt : 5.0 fs |
| dump interval(trj) : 100.0 fs |
| SHAKE algorithm : true (all bonds) |
| dump interval(Vbias) : 1.00 ps |
| Vbias prefactor (k) : 0.0338 Eh |
| Vbias exponent (α) : 0.2808 bohr⁻² |
*MTD 4 completed successfully ... 2 min, 0.400 sec
*MTD 7 completed successfully ... 2 min, 0.479 sec
*MTD 5 completed successfully ... 2 min, 0.911 sec
*MTD 8 completed successfully ... 2 min, 1.311 sec
*MTD 12 completed successfully ... 2 min, 1.473 sec
*MTD 2 completed successfully ... 2 min, 1.540 sec
*MTD 10 completed successfully ... 2 min, 1.569 sec
*MTD 11 completed successfully ... 2 min, 1.932 sec
*MTD 1 completed successfully ... 2 min, 2.180 sec
*MTD 6 completed successfully ... 2 min, 2.344 sec
*MTD 14 completed successfully ... 2 min, 2.349 sec
*MTD 9 completed successfully ... 2 min, 2.604 sec
*MTD 3 completed successfully ... 2 min, 2.632 sec
*MTD 13 completed successfully ... 2 min, 2.883 sec
======================================
| Multilevel Ensemble Optimization |
======================================
Optimizing all 1106 structures from file "crest_dynamics.trj" ...
----------------------
crude pre-optimization
----------------------
Optimization engine: ANCOPT
Hessian update type: BFGS
E/G convergence criteria: 0.500E-03 Eh, 0.100E-01 Eh/a0
maximum optimization steps: 200
|>0.1% |>7.5% |>15.0% |>22.5% |>30.0% |>37.5% |>45.0% |>52.5% |>60.0% |>67.5% |>75.0% |>82.5% |>90.1% |>97.6% |>100.0%
done.
> 1106 of 1106 structures successfully optimized (100.0% success)
> Total runtime for 1106 optimizations:
* wall-time: 0 d, 0 h, 1 min, 38.800 sec
* cpu-time: 0 d, 0 h, 22 min, 46.570 sec
* ratio c/w: 13.832 speedup
> Corresponding to approximately 0.089 sec per processed structure
CREGEN> running RMSDs ... done.
CREGEN> E lowest : -65.09606
776 structures remain within 12.00 kcal/mol window
----------------------------------
optimization with tight thresholds
----------------------------------
Optimization engine: ANCOPT
Hessian update type: BFGS
E/G convergence criteria: 0.100E-05 Eh, 0.800E-03 Eh/a0
maximum optimization steps: 200
|>0.1% |>10.1% |>20.1% |>30.0% |>40.1% |>50.0% |>60.1% |>70.1% |>80.0% |>90.1% |>100.0%
done.
> 776 of 776 structures successfully optimized (100.0% success)
> Total runtime for 776 optimizations:
* wall-time: 0 d, 0 h, 1 min, 34.821 sec
* cpu-time: 0 d, 0 h, 21 min, 46.152 sec
* ratio c/w: 13.775 speedup
> Corresponding to approximately 0.122 sec per processed structure
CREGEN> running RMSDs ... done.
CREGEN> E lowest : -65.09630
150 structures remain within 6.00 kcal/mol window
========================================
MTD Simulations done
========================================
Collecting ensmbles.
CREGEN> running RMSDs ... done.
CREGEN> E lowest : -65.09784
182 structures remain within 6.00 kcal/mol window
===============================================
Additional regular MDs on lowest 4 conformer(s)
===============================================
:::::::::::::: starting MD 1 ::::::::::::::
| MD simulation time : 4.0 ps |
| target T : 400.0 K |
| timestep dt : 5.0 fs |
| dump interval(trj) : 100.0 fs |
| SHAKE algorithm : true (all bonds) |
:::::::::::::: starting MD 8 ::::::::::::::
| MD simulation time : 4.0 ps |
| target T : 500.0 K |
| timestep dt : 5.0 fs |
| dump interval(trj) : 100.0 fs |
| SHAKE algorithm : true (all bonds) |
:::::::::::::: starting MD 6 ::::::::::::::
| MD simulation time : 4.0 ps |
| target T : 500.0 K |
| timestep dt : 5.0 fs |
| dump interval(trj) : 100.0 fs |
| SHAKE algorithm : true (all bonds) |
:::::::::::::: starting MD 2 ::::::::::::::
| MD simulation time : 4.0 ps |
| target T : 400.0 K |
| timestep dt : 5.0 fs |
| dump interval(trj) : 100.0 fs |
| SHAKE algorithm : true (all bonds) |
:::::::::::::: starting MD 3 ::::::::::::::
| MD simulation time : 4.0 ps |
| target T : 400.0 K |
| timestep dt : 5.0 fs |
| dump interval(trj) : 100.0 fs |
| SHAKE algorithm : true (all bonds) |
:::::::::::::: starting MD 5 ::::::::::::::
| MD simulation time : 4.0 ps |
| target T : 500.0 K |
| timestep dt : 5.0 fs |
| dump interval(trj) : 100.0 fs |
| SHAKE algorithm : true (all bonds) |
:::::::::::::: starting MD 4 ::::::::::::::
| MD simulation time : 4.0 ps |
| target T : 400.0 K |
| timestep dt : 5.0 fs |
| dump interval(trj) : 100.0 fs |
| SHAKE algorithm : true (all bonds) |
:::::::::::::: starting MD 7 ::::::::::::::
| MD simulation time : 4.0 ps |
| target T : 500.0 K |
| timestep dt : 5.0 fs |
| dump interval(trj) : 100.0 fs |
| SHAKE algorithm : true (all bonds) |
*MD 1 completed successfully ... 0 min, 54.351 sec
*MD 4 completed successfully ... 0 min, 54.539 sec
*MD 5 completed successfully ... 0 min, 54.938 sec
*MD 2 completed successfully ... 0 min, 54.984 sec
*MD 7 completed successfully ... 0 min, 54.993 sec
*MD 8 completed successfully ... 0 min, 55.006 sec
*MD 6 completed successfully ... 0 min, 55.034 sec
*MD 3 completed successfully ... 0 min, 55.062 sec
Appending file crest_rotamers_1.xyz with new structures
Optimizing all 494 structures from file "crest_rotamers_1.xyz" ...
----------------------------------
optimization with tight thresholds
----------------------------------
Optimization engine: ANCOPT
Hessian update type: BFGS
E/G convergence criteria: 0.100E-05 Eh, 0.800E-03 Eh/a0
maximum optimization steps: 200
|>0.2% |>10.1% |>20.0% |>30.2% |>40.1% |>50.0% |>60.1% |>70.0% |>80.2% |>90.1% |>100.0%
done.
> 494 of 494 structures successfully optimized (100.0% success)
> Total runtime for 494 optimizations:
* wall-time: 0 d, 0 h, 0 min, 45.615 sec
* cpu-time: 0 d, 0 h, 10 min, 19.974 sec
* ratio c/w: 13.592 speedup
> Corresponding to approximately 0.092 sec per processed structure
CREGEN> running RMSDs ... done.
CREGEN> E lowest : -65.09785
211 structures remain within 6.00 kcal/mol window
========================================
| Structure Crossing (GC) |
========================================
=============================
# threads = 14
=============================
input file name : crest_rotamers_2.xyz
number of atoms : 45
number of points on xyz files : 211
conformer energy window /kcal : 6.00
CN per atom difference cut-off : 0.3000
RMSD threshold (Ang, Bohr) : 0.2500 0.4724
max. # of generated structures : 400
# in E window 211
10.2 % done
35.2 % done
60.3 % done
finished.
average rmsd w.r.t input : 1.61661
number of clash discarded : 11478
removed identical structures : 9136
400 structures written to confcross.xyz
======================================
| Multilevel Ensemble Optimization |
======================================
Optimizing all 400 structures from file "confcross.xyz" ...
----------------------
crude pre-optimization
----------------------
Optimization engine: ANCOPT
Hessian update type: BFGS
E/G convergence criteria: 0.500E-03 Eh, 0.100E-01 Eh/a0
maximum optimization steps: 200
|>0.2% |>10.0% |>20.0% |>30.0% |>40.0% |>50.0% |>60.0% |>70.2% |>80.0% |>90.2% |>100.0%
done.
> 400 of 400 structures successfully optimized (100.0% success)
> Total runtime for 400 optimizations:
* wall-time: 0 d, 0 h, 0 min, 17.123 sec
* cpu-time: 0 d, 0 h, 3 min, 55.314 sec
* ratio c/w: 13.743 speedup
> Corresponding to approximately 0.043 sec per processed structure
CREGEN> running RMSDs ... done.
CREGEN> E lowest : -65.09782
96 structures remain within 12.00 kcal/mol window
----------------------------------
optimization with tight thresholds
----------------------------------
Optimization engine: ANCOPT
Hessian update type: BFGS
E/G convergence criteria: 0.100E-05 Eh, 0.800E-03 Eh/a0
maximum optimization steps: 200
|>1.0% |>10.4% |>20.8% |>30.2% |>40.6% |>50.0% |>60.4% |>70.8% |>80.2% |>90.6% |>100.0%
done.
> 96 of 96 structures successfully optimized (100.0% success)
> Total runtime for 96 optimizations:
* wall-time: 0 d, 0 h, 0 min, 6.321 sec
* cpu-time: 0 d, 0 h, 1 min, 23.505 sec
* ratio c/w: 13.211 speedup
> Corresponding to approximately 0.066 sec per processed structure
CREGEN> running RMSDs ... done.
CREGEN> E lowest : -65.09784
64 structures remain within 6.00 kcal/mol window
appending new structures to crest_rotamers_2.xyz
CREGEN> running RMSDs ... done.
CREGEN> E lowest : -65.09785
================================================
| Final Geometry Optimization |
================================================
Optimizing all 265 structures from file "crest_rotamers_3.xyz" ...
---------------------------------------
optimization with very tight thresholds
---------------------------------------
Optimization engine: ANCOPT
Hessian update type: BFGS
E/G convergence criteria: 0.100E-06 Eh, 0.200E-03 Eh/a0
maximum optimization steps: 200
|>0.4% |>10.2% |>20.0% |>30.2% |>40.0% |>50.2% |>60.0% |>70.2% |>80.0% |>90.2% |>100.0%
done.
> 265 of 265 structures successfully optimized (100.0% success)
> Total runtime for 265 optimizations:
* wall-time: 0 d, 0 h, 0 min, 8.178 sec
* cpu-time: 0 d, 0 h, 1 min, 50.809 sec
* ratio c/w: 13.550 speedup
> Corresponding to approximately 0.031 sec per processed structure
CREGEN> running RMSDs ... done.
CREGEN> E lowest : -65.09784
265 structures remain within 6.00 kcal/mol window
--------------------------
Final Ensemble Information
--------------------------
input file name : crest_rotamers_3.xyz
output file name : crest_rotamers_4.xyz
number of atoms : 45
number of points on xyz files : 265
RMSD threshold : 0.1250
Bconst threshold : 0.0100
population threshold : 0.0500
# fragment in coord : 1
# bonds in reference structure : 47
number of reliable points : 265
sorting energy window (EWIN) : 6.0000 / kcal*mol⁻¹
reference state Etot : -65.097837880000000
number of doubles removed by rot/RMSD : 0
total number unique points considered further : 265
Erel/kcal Etot weight/tot conformer set degen origin
1 0.000 -65.09784 0.05480 0.82181 1 15
2 0.000 -65.09784 0.05479
3 0.000 -65.09784 0.05479
4 0.000 -65.09784 0.05479
5 0.000 -65.09784 0.05479
6 0.000 -65.09784 0.05479
7 0.000 -65.09784 0.05479
8 0.000 -65.09784 0.05479
9 0.000 -65.09784 0.05479
10 0.000 -65.09784 0.05478
11 0.000 -65.09784 0.05478
12 0.000 -65.09784 0.05478
13 0.000 -65.09784 0.05478
14 0.000 -65.09784 0.05478
15 0.000 -65.09784 0.05477
16 0.966 -65.09630 0.01075 0.05373 2 5
17 0.966 -65.09630 0.01075
18 0.966 -65.09630 0.01075
19 0.966 -65.09630 0.01075
20 0.966 -65.09630 0.01074
21 1.300 -65.09577 0.00612 0.03058 3 5
22 1.300 -65.09577 0.00612
23 1.300 -65.09577 0.00612
24 1.300 -65.09577 0.00612
25 1.300 -65.09577 0.00612
26 1.741 -65.09506 0.00291 0.02906 4 10
27 1.741 -65.09506 0.00291
28 1.741 -65.09506 0.00291
29 1.741 -65.09506 0.00291
30 1.741 -65.09506 0.00291
31 1.741 -65.09506 0.00291
32 1.742 -65.09506 0.00291
33 1.742 -65.09506 0.00291
34 1.742 -65.09506 0.00291
35 1.742 -65.09506 0.00291
36 1.810 -65.09495 0.00259 0.03109 5 12
37 1.810 -65.09495 0.00259
38 1.810 -65.09495 0.00259
39 1.810 -65.09495 0.00259
40 1.810 -65.09495 0.00259
41 1.810 -65.09495 0.00259
42 1.810 -65.09495 0.00259
43 1.810 -65.09495 0.00259
44 1.810 -65.09495 0.00259
45 1.810 -65.09495 0.00259
46 1.810 -65.09495 0.00259
47 1.810 -65.09495 0.00259
48 1.871 -65.09486 0.00234 0.01401 6 6
49 1.871 -65.09486 0.00234
50 1.871 -65.09486 0.00234
51 1.871 -65.09486 0.00234
52 1.871 -65.09486 0.00234
53 1.871 -65.09486 0.00234
54 2.104 -65.09448 0.00158 0.00946 7 6
55 2.104 -65.09448 0.00158
56 2.104 -65.09448 0.00158
57 2.104 -65.09448 0.00158
58 2.104 -65.09448 0.00158
59 2.104 -65.09448 0.00158
60 2.197 -65.09434 0.00135 0.00539 8 4
61 2.197 -65.09434 0.00135
62 2.197 -65.09434 0.00135
63 2.197 -65.09434 0.00135
64 3.259 -65.09264 0.00022 0.00045 9 2
65 3.259 -65.09264 0.00022
66 3.348 -65.09250 0.00019 0.00058 10 3
67 3.348 -65.09250 0.00019
68 3.348 -65.09250 0.00019
69 3.729 -65.09190 0.00010 0.00020 11 2
70 3.729 -65.09190 0.00010
71 3.927 -65.09158 0.00007 0.00058 12 8
72 3.927 -65.09158 0.00007
73 3.927 -65.09158 0.00007
74 3.927 -65.09158 0.00007
75 3.927 -65.09158 0.00007
76 3.927 -65.09158 0.00007
77 3.927 -65.09158 0.00007
78 3.927 -65.09158 0.00007
79 4.363 -65.09089 0.00003 0.00035 13 10
80 4.363 -65.09089 0.00003
81 4.363 -65.09089 0.00003
82 4.363 -65.09089 0.00003
83 4.363 -65.09089 0.00003
84 4.363 -65.09089 0.00003
85 4.363 -65.09089 0.00003
86 4.363 -65.09088 0.00003
87 4.363 -65.09088 0.00003
88 4.363 -65.09088 0.00003
89 4.389 -65.09084 0.00003 0.00013 14 4
90 4.389 -65.09084 0.00003
91 4.389 -65.09084 0.00003
92 4.389 -65.09084 0.00003
93 4.424 -65.09079 0.00003 0.00060 15 19
94 4.424 -65.09079 0.00003
95 4.424 -65.09079 0.00003
96 4.424 -65.09079 0.00003
97 4.424 -65.09079 0.00003
98 4.424 -65.09079 0.00003
99 4.424 -65.09079 0.00003
100 4.424 -65.09079 0.00003
101 4.424 -65.09079 0.00003
102 4.424 -65.09079 0.00003
103 4.424 -65.09079 0.00003
104 4.424 -65.09079 0.00003
105 4.424 -65.09079 0.00003
106 4.424 -65.09079 0.00003
107 4.424 -65.09079 0.00003
108 4.424 -65.09079 0.00003
109 4.424 -65.09079 0.00003
110 4.424 -65.09079 0.00003
111 4.424 -65.09079 0.00003
112 4.514 -65.09064 0.00003 0.00073 16 27
113 4.514 -65.09064 0.00003
114 4.514 -65.09064 0.00003
115 4.514 -65.09064 0.00003
116 4.514 -65.09064 0.00003
117 4.514 -65.09064 0.00003
118 4.514 -65.09064 0.00003
119 4.514 -65.09064 0.00003
120 4.514 -65.09064 0.00003
121 4.514 -65.09064 0.00003
122 4.514 -65.09064 0.00003
123 4.514 -65.09064 0.00003
124 4.514 -65.09064 0.00003
125 4.514 -65.09064 0.00003
126 4.514 -65.09064 0.00003
127 4.514 -65.09064 0.00003
128 4.514 -65.09064 0.00003
129 4.514 -65.09064 0.00003
130 4.514 -65.09064 0.00003
131 4.514 -65.09064 0.00003
132 4.514 -65.09064 0.00003
133 4.514 -65.09064 0.00003
134 4.514 -65.09064 0.00003
135 4.514 -65.09064 0.00003
136 4.514 -65.09064 0.00003
137 4.514 -65.09064 0.00003
138 4.514 -65.09064 0.00003
139 4.540 -65.09060 0.00003 0.00054 17 21
140 4.540 -65.09060 0.00003
141 4.540 -65.09060 0.00003
142 4.540 -65.09060 0.00003
143 4.540 -65.09060 0.00003
144 4.540 -65.09060 0.00003
145 4.540 -65.09060 0.00003
146 4.540 -65.09060 0.00003
147 4.540 -65.09060 0.00003
148 4.540 -65.09060 0.00003
149 4.540 -65.09060 0.00003
150 4.540 -65.09060 0.00003
151 4.540 -65.09060 0.00003
152 4.540 -65.09060 0.00003
153 4.540 -65.09060 0.00003
154 4.540 -65.09060 0.00003
155 4.540 -65.09060 0.00003
156 4.540 -65.09060 0.00003
157 4.540 -65.09060 0.00003
158 4.540 -65.09060 0.00003
159 4.540 -65.09060 0.00003
160 4.682 -65.09038 0.00002 0.00004 18 2
161 4.682 -65.09038 0.00002
162 4.785 -65.09021 0.00002 0.00002 19 1
163 4.892 -65.09004 0.00001 0.00001 20 1
164 4.927 -65.08999 0.00001 0.00015 21 11
165 4.927 -65.08999 0.00001
166 4.927 -65.08999 0.00001
167 4.927 -65.08999 0.00001
168 4.927 -65.08999 0.00001
169 4.927 -65.08999 0.00001
170 4.927 -65.08999 0.00001
171 4.928 -65.08999 0.00001
172 4.928 -65.08999 0.00001
173 4.928 -65.08999 0.00001
174 4.928 -65.08999 0.00001
175 4.966 -65.08992 0.00001 0.00001 22 1
176 5.041 -65.08980 0.00001 0.00004 23 4
177 5.041 -65.08980 0.00001
178 5.041 -65.08980 0.00001
179 5.041 -65.08980 0.00001
180 5.097 -65.08971 0.00001 0.00002 24 2
181 5.097 -65.08971 0.00001
182 5.101 -65.08971 0.00001 0.00002 25 2
183 5.101 -65.08971 0.00001
184 5.193 -65.08956 0.00001 0.00011 26 13
185 5.193 -65.08956 0.00001
186 5.193 -65.08956 0.00001
187 5.193 -65.08956 0.00001
188 5.193 -65.08956 0.00001
189 5.193 -65.08956 0.00001
190 5.193 -65.08956 0.00001
191 5.193 -65.08956 0.00001
192 5.193 -65.08956 0.00001
193 5.193 -65.08956 0.00001
194 5.193 -65.08956 0.00001
195 5.193 -65.08956 0.00001
196 5.193 -65.08956 0.00001
197 5.333 -65.08934 0.00001 0.00001 27 1
198 5.520 -65.08904 0.00000 0.00000 28 1
199 5.536 -65.08902 0.00000 0.00001 29 2
200 5.536 -65.08902 0.00000
201 5.538 -65.08901 0.00000 0.00011 30 23
202 5.538 -65.08901 0.00000
203 5.538 -65.08901 0.00000
204 5.538 -65.08901 0.00000
205 5.538 -65.08901 0.00000
206 5.538 -65.08901 0.00000
207 5.538 -65.08901 0.00000
208 5.538 -65.08901 0.00000
209 5.538 -65.08901 0.00000
210 5.538 -65.08901 0.00000
211 5.538 -65.08901 0.00000
212 5.538 -65.08901 0.00000
213 5.538 -65.08901 0.00000
214 5.538 -65.08901 0.00000
215 5.538 -65.08901 0.00000
216 5.538 -65.08901 0.00000
217 5.538 -65.08901 0.00000
218 5.538 -65.08901 0.00000
219 5.538 -65.08901 0.00000
220 5.538 -65.08901 0.00000
221 5.538 -65.08901 0.00000
222 5.538 -65.08901 0.00000
223 5.538 -65.08901 0.00000
224 5.642 -65.08885 0.00000 0.00000 31 1
225 5.802 -65.08859 0.00000 0.00006 32 21
226 5.802 -65.08859 0.00000
227 5.802 -65.08859 0.00000
228 5.802 -65.08859 0.00000
229 5.802 -65.08859 0.00000
230 5.802 -65.08859 0.00000
231 5.802 -65.08859 0.00000
232 5.802 -65.08859 0.00000
233 5.802 -65.08859 0.00000
234 5.802 -65.08859 0.00000
235 5.802 -65.08859 0.00000
236 5.802 -65.08859 0.00000
237 5.802 -65.08859 0.00000
238 5.802 -65.08859 0.00000
239 5.802 -65.08859 0.00000
240 5.802 -65.08859 0.00000
241 5.802 -65.08859 0.00000
242 5.802 -65.08859 0.00000
243 5.802 -65.08859 0.00000
244 5.802 -65.08859 0.00000
245 5.802 -65.08859 0.00000
246 5.881 -65.08847 0.00000 0.00005 33 20
247 5.881 -65.08847 0.00000
248 5.881 -65.08847 0.00000
249 5.881 -65.08847 0.00000
250 5.881 -65.08847 0.00000
251 5.881 -65.08847 0.00000
252 5.881 -65.08847 0.00000
253 5.881 -65.08847 0.00000
254 5.881 -65.08847 0.00000
255 5.881 -65.08847 0.00000
256 5.881 -65.08847 0.00000
257 5.881 -65.08847 0.00000
258 5.881 -65.08847 0.00000
259 5.881 -65.08847 0.00000
260 5.881 -65.08847 0.00000
261 5.881 -65.08847 0.00000
262 5.881 -65.08847 0.00000
263 5.881 -65.08847 0.00000
264 5.881 -65.08847 0.00000
265 5.881 -65.08847 0.00000
T /K : 298.15
E lowest : -65.09784
ensemble average energy (kcal) : 0.277
ensemble entropy (J/mol K, cal/mol K) : 28.033 6.700
ensemble free energy (kcal/mol) : -1.998
population of lowest in % : 82.181
number of unique conformers for further calc 33
list of relative energies saved as "crest.energies"
-----------------
Wall Time Summary
-----------------
CREST runtime (total) 0 d, 0 h, 12 min, 54.649 sec
------------------------------------------------------------------
Trial metadynamics (MTD) ... 0 min, 11.726 sec ( 1.514%)
Metadynamics (MTD) ... 4 min, 7.803 sec ( 31.989%)
Geometry optimization ... 7 min, 14.403 sec ( 56.077%)
Molecular dynamics (MD) ... 0 min, 55.106 sec ( 7.114%)
Genetic crossing (GC) ... 0 min, 24.307 sec ( 3.138%)
I/O and setup ... 0 min, 1.304 sec ( 0.168%)
------------------------------------------------------------------
* wall-time: 0 d, 0 h, 12 min, 54.649 sec
* cpu-time: 0 d, 2 h, 49 min, 10.989 sec
* ratio c/w: 13.104 speedup
------------------------------------------------------------------
* Total number of energy+grad calls: 148615
CREST terminated normally.
結果
出力されている crest_conformers.xyz をUCSF ChimeraXで開いて構造を確認しました。
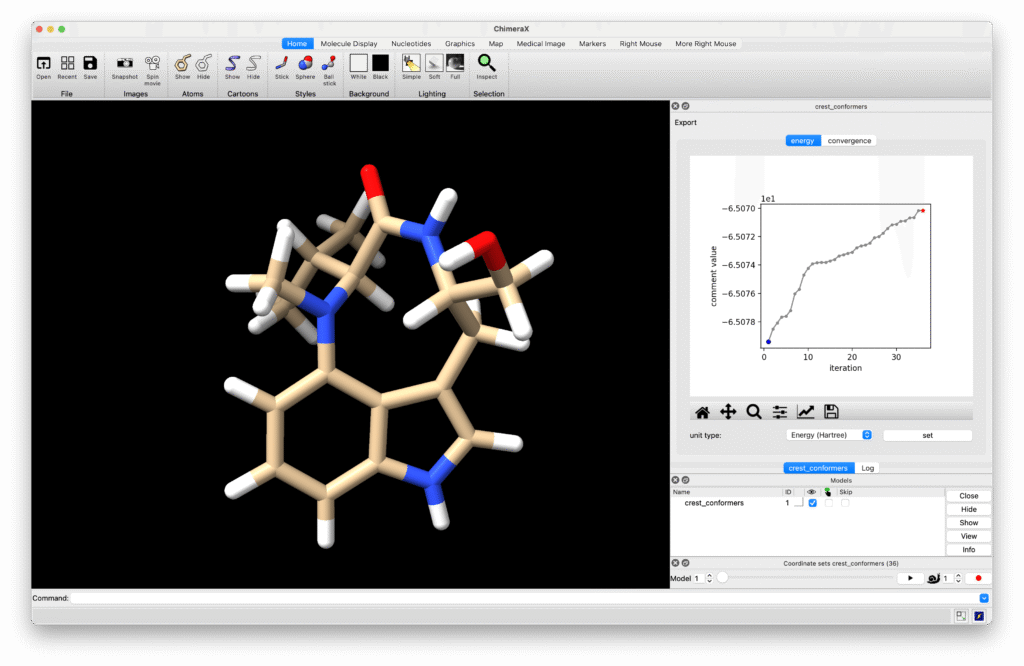
33種類のコンホマーが含まれており、cis-amide型のtwist、foldコンホマー、trans-amide型のsofaコンホマーが確認できました。NMRでは観測されないfoldコンホマーが出てきていましたが、より正確な計算はGaussianなりORCAなりで行えばよいので、配座探索としては計算時間、精度ともに十分だと考えられます。RDKitを使った配座探索ではtrans-amide型が出現しなかったので、MDを基にした配座探索が少くともこの手の化合物には適していることが確認できました。
(了)
コメント失礼します。香大農渡邉研の院生です。
Alphafoldで検索してたら柳田先生のブログが見つかってびっくりしました。
色々便利なツールや統計学的検証を分かり易くまとめられていて今使うかはさておきとても参考になりました。
これからも研究の益々のご発展をお祈り申し上げます。
コメントをいただきありがとうございます。AlphaFoldに関する記事を書いたのはかなり前で、専門家でもないのであまり役に立つ内容ではなかったかと。研究生活を楽しんでください。