目的: 受容体の一次構造からリガンド親和性を予測するモデルを作る.
環境: R 3.3.2 GUI 1.68 Mavericks build (7288).
参考サイト: “R”: Predicting a Test Set (Gasoline)
ケモメトリクス(化学計量学)の手法について実習したのでメモ.
定量的配列活性モデル(Quantitative sequence-activity model,QSAM)は,比較的短いDNA,RNA,ペプチドの定量的構造活性相関(QSAR)で使用される手法ですが,今回は中程度のタンパク質ドメインに対して適用してみることにしました.
Contents
統計ソフトRのインストール
$ brew tap caskroom/cask
$ brew cask install r-appGUI版をインストールしました.
トレーニングセット
C1ドメインの一次構造(アミノ酸配列,PKCはラットの配列)とリガンド ([3H]phorbol-12,13-dibutyrate) の解離定数Kd.
Irie(1999), Shindo(2003), Irie(2004) から解離定数Kdのデータを抽出しました.
PKCa-C1A HKFIARFFKQPTFCSHCTDFIWG-FGKQGFQCQVCCFVVHKRCHEFVTFSC
PKCb-C1A HKFTARFFKQPTFCSHCTDFIWG-FGKQGFQCQVCCFVVHKRCHEFVTFSC
PKCg-C1A HKFTARFFKQPTFCSHCTDFIWG-IGKQGLQCQVCSFVVHRRCHEFVTFEC
PKCd-C1A HEFIATFFGQPTFCSVCKEFVWG-LNKQGYKCRQCNAAIHKKCIDKIIGRC
PKCe-C1A HKFMATYLRQPTYCSHCRDFIWGVIGKQGYQCQVCTCVVHKRCHELIITKC
PKCh-C1A HKFMATYLRQPTYCSHCREFIWGVFGKQGYQCQVCTCVVHKRCHHLIVTAC
PKCq-C1A HEFTATFFPQPTFCSVCHEFVWG-LNKQGYQCRRCNAAIHKKCIDKVIAKC
PKCa-C1B HKFKIHTYGSPTFCDHCGSLLYG-LIHQGMKCDTCDMNVHKQCVINVPSLC
PKCb-C1B HKFKIHTYSSPTFCDHCGSLLYG-LIHQGMKCDTCMMNVHKRCVMNVPSLC
PKCg-C1B HKFRLHSYSSPTFCDHCGSLLYG-LVHQGMKCSCCEMNVHRRCVRSVPSLC
PKCd-C1B HRFKVYNYMSPTFCDHCGTLLWG-LVKQGLKCEDCGMNVHHKCREKVANLC
PKCe-C1B HKFGIHNYKVPTFCDHCGSLLWG-LLRQGLQCKVCKMNVHRRCETNVAPNC
PKCh-C1B HKFNVHNYKVPTFCDHCGSLLWG-IMRQGLQCKICKMNVHIRCQANVAPNC
PKCq-C1B HRFKVYNYKSPTFCEHCGTLLWG-LARQGLKCDACGMNVHHRCQTKVANLC
RasGRP1 HNFQETTYLKPTFCDNCAGFLWG-VIKQGYRCKDCGMNCHKQCKDLVVFEC
RasGRP2 HNFQESNSLRPVACRHCKALILG-IYKQGLKCRACGVNCHKQCKDRLSVEC
RasGRP3 HNFQEMTYLKPTFCEHCAGFLWG-IIKQGYKCKDCGANCHKQCKDLLVLAC
RasGRP4 HTFHEVTFRKPTFCDSCSGFLWG-VTKQGYRCRECGLCCHKHCRDQVKVEC
a-chimaerin HNFKVHTFRGPHWCEYCANFMWG-LIAQGVKCADCGLNVHKQCSKMVPNDC
b-chimaerin HNFKVHTFRGPHWCEYCANFMWG-LIAQGVRCSDCGLNVHKQCSKHVPNDC
Unc13(C.elegans) HNFATTTFQTPTFCYECEGLLWG-LARQGLRCTQCQVKVHDKCRELLSADC
Munc13(human) HNFEVWTATTPTYCYECEGLLWG-IARQGMRCTECGVKCHEKCQDLLNADC
PKD-C1A HALFVHSYRAPAFCDHCGEMLWG-LVRQGLKCEGCGLNYHKRCAFKIPNNC
PKD-C1B HTFVIHSYTRPTVCQYCKKLLKG-LFRQGLQCKDCRFNCHKRCAPKVPNNC
DGKb-C1A(rat) HVWRLKHFNKPAYCNLCLNMLIG-VGKQGLCCSFCKYTVHERCA-RAPPSC
DGKb-C1A(human) HVWRLKHFNKPAYCNLCLNMLIG-VGKQGLCCSFCKYTVHERCVARAPPSC
DGKg-C1A HAWTMKHFKKPTYCNFCHIMLMG-VRKQGLCCTYCKYTVHERCVSRNIPGC
* * * * :: * . ** * * * :* *
アミノ酸配列はそのままでは定量的に扱うことはできないので,何らかの数値(記述子, descriptor)でアミノ酸残基を表わす必要があります.これまでに様々なアミノ酸記述子が考案されていますが (Qian, 2017),今回は,Collantes and Dunn III(1995) によるアミノ酸記述子,すなわち等方性表面積(isotropic surface area, ISA)と電荷指数(electronic charge index, ESI)を使用しました.ISAとESIはどちらも正の値で,それぞれアミノ酸側鎖の嵩高さと局所的な極性を表わしています.例えば,グリシンはISA = 19.93, ECI = 0.02,フェニルアラニンはISA = 189.42, ECI = 0.14,アルギニンはISA = 52.98,ECI = 1.69となっています.
まず,Excelで表を作りCSV形式で書き出します.マクロを一回書くと以後効率的です.pKd = −log Kdです.以下のファイルをc1.csvとして保存します.アラインメントした時のギャップにはISA,ECI共に値0を入れました.
peptide,pKd,ISA1,ISA2,ISA3,ISA4,ISA5,ISA6,ISA7,ISA8,ISA9,ISA10,ISA11,ISA12,ISA13,ISA14,ISA15,ISA16,ISA17,ISA18,ISA19,ISA20,ISA21,ISA22,ISA23,ISA24,ISA25,ISA26,ISA27,ISA28,ISA29,ISA30,ISA31,ISA32,ISA33,ISA34,ISA35,ISA36,ISA37,ISA38,ISA39,ISA40,ISA41,ISA42,ISA43,ISA44,ISA45,ISA46,ISA47,ISA48,ISA49,ISA50,ISA51,ECI1,ECI2,ECI3,ECI4,ECI5,ECI6,ECI7,ECI8,ECI9,ECI10,ECI11,ECI12,ECI13,ECI14,ECI15,ECI16,ECI17,ECI18,ECI19,ECI20,ECI21,ECI22,ECI23,ECI24,ECI25,ECI26,ECI27,ECI28,ECI29,ECI30,ECI31,ECI32,ECI33,ECI34,ECI35,ECI36,ECI37,ECI38,ECI39,ECI40,ECI41,ECI42,ECI43,ECI44,ECI45,ECI46,ECI47,ECI48,ECI49,ECI50,ECI51 a-C1A,8.96,87.38,102.8,189.4,149.8,62.9,52.98,189.4,189.4,102.8,19.53,122.4,59.44,189.4,78.51,19.75,87.38,78.51,59.44,18.46,189.4,149.8,179.2,19.93,0,189.4,19.93,102.8,19.53,19.93,189.4,19.53,78.51,19.53,120.9,78.51,78.51,189.4,120.9,120.9,87.38,102.8,52.98,78.51,87.38,30.19,189.4,120.9,59.44,189.4,19.75,78.51,0.56,0.53,0.14,0.09,0.05,1.69,0.14,0.14,0.53,1.36,0.16,0.65,0.14,0.15,0.56,0.56,0.15,0.65,1.25,0.14,0.09,1.08,0.02,0,0.14,0.02,0.53,1.36,0.02,0.14,1.36,0.15,1.36,0.07,0.15,0.15,0.14,0.07,0.07,0.56,0.53,1.69,0.15,0.56,1.31,0.14,0.07,0.65,0.14,0.56,0.15 b-C1A,8.89,87.38,102.8,189.4,59.44,62.9,52.98,189.4,189.4,102.8,19.53,122.4,59.44,189.4,78.51,19.75,87.38,78.51,59.44,18.46,189.4,149.8,179.2,19.93,0,189.4,19.93,102.8,19.53,19.93,189.4,19.53,78.51,19.53,120.9,78.51,78.51,189.4,120.9,120.9,87.38,102.8,52.98,78.51,87.38,30.19,189.4,120.9,59.44,189.4,19.75,78.51,0.56,0.53,0.14,0.65,0.05,1.69,0.14,0.14,0.53,1.36,0.16,0.65,0.14,0.15,0.56,0.56,0.15,0.65,1.25,0.14,0.09,1.08,0.02,0,0.14,0.02,0.53,1.36,0.02,0.14,1.36,0.15,1.36,0.07,0.15,0.15,0.14,0.07,0.07,0.56,0.53,1.69,0.15,0.56,1.31,0.14,0.07,0.65,0.14,0.56,0.15 g-C1A,8.82,87.38,102.8,189.4,59.44,62.9,52.98,189.4,189.4,102.8,19.53,122.4,59.44,189.4,78.51,19.75,87.38,78.51,59.44,18.46,189.4,149.8,179.2,19.93,0,149.8,19.93,102.8,19.53,19.93,154.4,19.53,78.51,19.53,120.9,78.51,19.75,189.4,120.9,120.9,87.38,52.98,52.98,78.51,87.38,30.19,189.4,120.9,59.44,189.4,30.19,78.51,0.56,0.53,0.14,0.65,0.05,1.69,0.14,0.14,0.53,1.36,0.16,0.65,0.14,0.15,0.56,0.56,0.15,0.65,1.25,0.14,0.09,1.08,0.02,0,0.09,0.02,0.53,1.36,0.02,0.1,1.36,0.15,1.36,0.07,0.15,0.56,0.14,0.07,0.07,0.56,1.69,1.69,0.15,0.56,1.31,0.14,0.07,0.65,0.14,1.31,0.15 d-C1A,7.28,87.38,30.19,189.4,59.44,62.9,59.44,189.4,189.4,19.93,19.53,122.4,59.44,189.4,78.51,19.75,120.9,78.51,102.8,30.19,189.4,120.9,179.2,19.93,0,154.4,17.87,102.8,19.53,19.93,132.2,102.8,78.51,52.98,19.53,78.51,17.87,62.9,62.9,149.8,87.38,102.8,102.8,78.51,149.8,18.46,102.8,149.8,149.8,19.93,52.98,78.51,0.56,1.31,0.14,0.65,0.05,0.65,0.14,0.14,0.02,1.36,0.16,0.65,0.14,0.15,0.56,0.07,0.15,0.53,1.31,0.14,0.07,1.08,0.02,0,0.1,1.31,0.53,1.36,0.02,0.72,0.53,0.15,1.69,1.36,0.15,1.31,0.05,0.05,0.09,0.56,0.53,0.53,0.15,0.09,1.25,0.53,0.09,0.09,0.02,1.69,0.15 e-C1A,8.25,87.38,102.8,189.4,132.2,62.9,59.44,132.2,154.4,52.98,19.53,122.4,59.44,132.2,78.51,19.75,87.38,78.51,52.98,18.46,189.4,149.8,179.2,19.93,120.9,149.8,19.93,102.8,19.53,19.93,132.2,19.53,78.51,19.53,120.9,78.51,59.44,78.51,120.9,120.9,87.38,102.8,52.98,78.51,87.38,30.19,154.4,149.8,149.8,59.44,102.8,78.51,0.56,0.53,0.14,0.34,0.05,0.65,0.72,0.1,1.69,1.36,0.16,0.65,0.72,0.15,0.56,0.56,0.15,1.69,1.25,0.14,0.09,1.08,0.02,0.07,0.09,0.02,0.53,1.36,0.02,0.72,1.36,0.15,1.36,0.07,0.15,0.65,0.15,0.07,0.07,0.56,0.53,1.69,0.15,0.56,1.31,0.1,0.09,0.09,0.65,0.53,0.15 h-C1A,8.37,87.38,102.8,189.4,132.2,62.9,59.44,132.2,154.4,52.98,19.53,122.4,59.44,132.2,78.51,19.75,87.38,78.51,52.98,30.19,189.4,149.8,179.2,19.93,120.9,189.4,19.93,102.8,19.53,19.93,132.2,19.53,78.51,19.53,120.9,78.51,59.44,78.51,120.9,120.9,87.38,102.8,52.98,78.51,87.38,87.38,154.4,149.8,120.9,59.44,62.9,78.51,0.56,0.53,0.14,0.34,0.05,0.65,0.72,0.1,1.69,1.36,0.16,0.65,0.72,0.15,0.56,0.56,0.15,1.69,1.31,0.14,0.09,1.08,0.02,0.07,0.14,0.02,0.53,1.36,0.02,0.72,1.36,0.15,1.36,0.07,0.15,0.65,0.15,0.07,0.07,0.56,0.53,1.69,0.15,0.56,0.56,0.1,0.09,0.07,0.65,0.05,0.15 q-C1A,6.70,87.38,30.19,189.4,59.44,62.9,59.44,189.4,189.4,122.4,19.53,122.4,59.44,189.4,78.51,19.75,120.9,78.51,87.38,30.19,189.4,120.9,179.2,19.93,0,154.4,17.87,102.8,19.53,19.93,132.2,19.53,78.51,52.98,19.53,78.51,17.87,62.9,62.9,149.8,87.38,102.8,102.8,78.51,149.8,18.46,102.8,120.9,149.8,62.9,102.8,78.51,0.56,1.31,0.14,0.65,0.05,0.65,0.14,0.14,0.16,1.36,0.16,0.65,0.14,0.15,0.56,0.07,0.15,0.56,1.31,0.14,0.07,1.08,0.02,0,0.1,1.31,0.53,1.36,0.02,0.72,1.36,0.15,1.69,1.36,0.15,1.31,0.05,0.05,0.09,0.56,0.53,0.53,0.15,0.09,1.25,0.53,0.07,0.09,0.05,0.53,0.15 a-C1B,8.28,87.38,102.8,189.4,102.8,149.8,87.38,59.44,132.2,19.93,19.75,122.4,59.44,189.4,78.51,18.46,87.38,78.51,19.93,19.75,154.4,154.4,132.2,19.93,0,154.4,149.8,87.38,19.53,19.93,132.2,102.8,78.51,18.46,59.44,78.51,18.46,132.2,17.87,120.9,87.38,102.8,19.53,78.51,120.9,149.8,17.87,120.9,122.4,19.75,154.4,78.51,0.56,0.53,0.14,0.53,0.09,0.56,0.65,0.72,0.02,0.56,0.16,0.65,0.14,0.15,1.25,0.56,0.15,0.02,0.56,0.1,0.1,0.72,0.02,0,0.1,0.09,0.56,1.36,0.02,0.34,0.53,0.15,1.25,0.65,0.15,1.25,0.34,1.31,0.07,0.56,0.53,1.36,0.15,0.07,0.09,1.31,0.07,0.16,0.56,0.1,0.15 b-C1B,8.89,87.38,102.8,189.4,102.8,149.8,87.38,59.44,132.2,19.75,19.75,122.4,59.44,189.4,78.51,18.46,87.38,78.51,19.93,19.75,154.4,154.4,132.2,19.93,0,154.4,149.8,87.38,19.53,19.93,132.2,102.8,78.51,18.46,59.44,78.51,132.2,132.2,17.87,120.9,87.38,102.8,52.98,78.51,120.9,132.2,17.87,120.9,122.4,19.75,154.4,78.51,0.56,0.53,0.14,0.53,0.09,0.56,0.65,0.72,0.56,0.56,0.16,0.65,0.14,0.15,1.25,0.56,0.15,0.02,0.56,0.1,0.1,0.72,0.02,0,0.1,0.09,0.56,1.36,0.02,0.34,0.53,0.15,1.25,0.65,0.15,0.34,0.34,1.31,0.07,0.56,0.53,1.69,0.15,0.07,0.34,1.31,0.07,0.16,0.56,0.1,0.15 g-C1B,8.92,87.38,102.8,189.4,52.98,154.4,87.38,19.75,132.2,19.75,19.75,122.4,59.44,189.4,78.51,18.46,87.38,78.51,19.93,19.75,154.4,154.4,132.2,19.93,0,154.4,120.9,87.38,19.53,19.93,132.2,102.8,78.51,19.75,78.51,78.51,30.19,132.2,17.87,120.9,87.38,52.98,52.98,78.51,120.9,52.98,19.75,120.9,122.4,19.75,154.4,78.51,0.56,0.53,0.14,1.69,0.1,0.56,0.56,0.72,0.56,0.56,0.16,0.65,0.14,0.15,1.25,0.56,0.15,0.02,0.56,0.1,0.1,0.72,0.02,0,0.1,0.07,0.56,1.36,0.02,0.34,0.53,0.15,0.56,0.15,0.15,1.31,0.34,1.31,0.07,0.56,1.69,1.69,0.15,0.07,1.69,0.56,0.07,0.16,0.56,0.1,0.15 d-C1B,9.28,87.38,52.98,189.4,102.8,120.9,132.2,17.87,132.2,132.2,19.75,122.4,59.44,189.4,78.51,18.46,87.38,78.51,19.93,19.75,154.4,154.4,179.2,19.93,0,154.4,120.9,102.8,19.53,19.93,154.4,102.8,78.51,30.19,18.46,78.51,19.93,132.2,17.87,120.9,87.38,87.38,102.8,78.51,52.98,30.19,102.8,120.9,62.9,17.87,154.4,78.51,0.56,1.69,0.14,0.53,0.07,0.72,1.31,0.72,0.34,0.56,0.16,0.65,0.14,0.15,1.25,0.56,0.15,0.02,0.56,0.1,0.1,1.08,0.02,0,0.1,0.07,0.53,1.36,0.02,0.1,0.53,0.15,1.31,1.25,0.15,0.02,0.34,1.31,0.07,0.56,0.56,0.53,0.15,1.69,1.31,0.53,0.07,0.05,1.31,0.1,0.15 e-C1B,9.09,87.38,102.8,189.4,19.93,149.8,87.38,17.87,132.2,102.8,120.9,122.4,59.44,189.4,78.51,18.46,87.38,78.51,19.93,19.75,154.4,154.4,179.2,19.93,0,154.4,154.4,52.98,19.53,19.93,154.4,19.53,78.51,102.8,120.9,78.51,102.8,132.2,17.87,120.9,87.38,52.98,52.98,78.51,30.19,59.44,17.87,120.9,62.9,122.4,17.87,78.51,0.56,0.53,0.14,0.02,0.09,0.56,1.31,0.72,0.53,0.07,0.16,0.65,0.14,0.15,1.25,0.56,0.15,0.02,0.56,0.1,0.1,1.08,0.02,0,0.1,0.1,1.69,1.36,0.02,0.1,1.36,0.15,0.53,0.07,0.15,0.53,0.34,1.31,0.07,0.56,1.69,1.69,0.15,1.31,0.65,1.31,0.07,0.05,0.16,1.31,0.15 h-C1B,9.35,87.38,102.8,189.4,17.87,120.9,87.38,17.87,132.2,102.8,120.9,122.4,59.44,189.4,78.51,18.46,87.38,78.51,19.93,19.75,154.4,154.4,179.2,19.93,0,149.8,132.2,52.98,19.53,19.93,154.4,19.53,78.51,102.8,149.8,78.51,102.8,132.2,17.87,120.9,87.38,149.8,52.98,78.51,19.53,62.9,17.87,120.9,62.9,122.4,17.87,78.51,0.56,0.53,0.14,1.31,0.07,0.56,1.31,0.72,0.53,0.07,0.16,0.65,0.14,0.15,1.25,0.56,0.15,0.02,0.56,0.1,0.1,1.08,0.02,0,0.09,0.34,1.69,1.36,0.02,0.1,1.36,0.15,0.53,0.09,0.15,0.53,0.34,1.31,0.07,0.56,0.09,1.69,0.15,1.36,0.05,1.31,0.07,0.05,0.16,1.31,0.15 q-C1B,9.14,87.38,52.98,189.4,102.8,120.9,132.2,17.87,132.2,102.8,19.75,122.4,59.44,189.4,78.51,30.19,87.38,78.51,19.93,59.44,154.4,154.4,179.2,19.93,0,154.4,62.9,52.98,19.53,19.93,154.4,102.8,78.51,18.46,62.9,78.51,19.93,132.2,17.87,120.9,87.38,87.38,52.98,78.51,19.53,59.44,102.8,120.9,62.9,17.87,154.4,78.51,0.56,1.69,0.14,0.53,0.07,0.72,1.31,0.72,0.53,0.56,0.16,0.65,0.14,0.15,1.31,0.56,0.15,0.02,0.65,0.1,0.1,1.08,0.02,0,0.1,0.05,1.69,1.36,0.02,0.1,0.53,0.15,1.25,0.05,0.15,0.02,0.34,1.31,0.07,0.56,0.56,1.69,0.15,1.36,0.65,0.53,0.07,0.05,1.31,0.1,0.15 RasGRP1,9.14,87.38,17.87,189.4,19.53,30.19,59.44,59.44,132.2,154.4,102.8,122.4,59.44,189.4,78.51,18.46,17.87,78.51,62.9,19.93,189.4,154.4,179.2,19.93,0,120.9,149.8,102.8,19.53,19.93,132.2,52.98,78.51,102.8,18.46,78.51,19.93,132.2,17.87,78.51,87.38,102.8,19.53,78.51,102.8,18.46,154.4,120.9,120.9,189.4,30.19,78.51,0.56,1.31,0.14,1.36,1.31,0.65,0.65,0.72,0.1,0.53,0.16,0.65,0.14,0.15,1.25,1.31,0.15,0.05,0.02,0.14,0.1,1.08,0.02,0,0.07,0.09,0.53,1.36,0.02,0.72,1.69,0.15,0.53,1.25,0.15,0.02,0.34,1.31,0.15,0.56,0.53,1.36,0.15,0.53,1.25,0.1,0.07,0.07,0.14,1.31,0.15 RasGRP2,6.00,87.38,17.87,189.4,19.53,30.19,19.75,17.87,19.75,154.4,52.98,122.4,120.9,62.9,78.51,52.98,87.38,78.51,102.8,62.9,154.4,149.8,154.4,19.93,0,149.8,132.2,102.8,19.53,19.93,154.4,102.8,78.51,52.98,62.9,78.51,19.93,120.9,17.87,78.51,87.38,102.8,19.53,78.51,102.8,18.46,52.98,154.4,19.75,120.9,30.19,78.51,0.56,1.31,0.14,1.36,1.31,0.56,1.31,0.56,0.1,1.69,0.16,0.07,0.05,0.15,1.69,0.56,0.15,0.53,0.05,0.1,0.09,0.1,0.02,0,0.09,0.72,0.53,1.36,0.02,0.1,0.53,0.15,1.69,0.05,0.15,0.02,0.07,1.31,0.15,0.56,0.53,1.36,0.15,0.53,1.25,1.69,0.1,0.56,0.07,1.31,0.15 RasGRP3,8.82,87.38,17.87,189.4,19.53,30.19,132.2,59.44,132.2,154.4,102.8,122.4,59.44,189.4,78.51,30.19,87.38,78.51,62.9,19.93,189.4,154.4,179.2,19.93,0,149.8,149.8,102.8,19.53,19.93,132.2,102.8,78.51,102.8,18.46,78.51,19.93,62.9,17.87,78.51,87.38,102.8,19.53,78.51,102.8,18.46,154.4,154.4,120.9,154.4,62.9,78.51,0.56,1.31,0.14,1.36,1.31,0.34,0.65,0.72,0.1,0.53,0.16,0.65,0.14,0.15,1.31,0.56,0.15,0.05,0.02,0.14,0.1,1.08,0.02,0,0.09,0.09,0.53,1.36,0.02,0.72,0.53,0.15,0.53,1.25,0.15,0.02,0.05,1.31,0.15,0.56,0.53,1.36,0.15,0.53,1.25,0.1,0.1,0.07,0.1,0.05,0.15 RasGRP4,8.97,87.38,59.44,189.4,87.38,30.19,120.9,59.44,189.4,52.98,102.8,122.4,59.44,189.4,78.51,18.46,19.75,78.51,19.75,19.93,189.4,154.4,179.2,19.93,0,120.9,59.44,102.8,19.53,19.93,132.2,52.98,78.51,52.98,30.19,78.51,19.93,154.4,78.51,78.51,87.38,102.8,87.38,78.51,52.98,18.46,19.53,120.9,102.8,120.9,30.19,78.51,0.56,0.65,0.14,0.56,1.31,0.07,0.65,0.14,1.69,0.53,0.16,0.65,0.14,0.15,1.25,0.56,0.15,0.56,0.02,0.14,0.1,1.08,0.02,0,0.07,0.65,0.53,1.36,0.02,0.72,1.69,0.15,1.69,1.31,0.15,0.02,0.1,0.15,0.15,0.56,0.53,0.56,0.15,1.69,1.25,1.36,0.07,0.53,0.07,1.31,0.15 a-chimaerin,8.32,87.38,17.87,189.4,102.8,120.9,87.38,59.44,189.4,52.98,19.93,122.4,87.38,179.2,78.51,30.19,132.2,78.51,62.9,17.87,189.4,132.2,179.2,19.93,0,154.4,149.8,62.9,19.53,19.93,120.9,102.8,78.51,62.9,18.46,78.51,19.93,154.4,17.87,120.9,87.38,102.8,19.53,78.51,19.75,102.8,132.2,120.9,122.4,17.87,18.46,78.51,0.56,1.31,0.14,0.53,0.07,0.56,0.65,0.14,1.69,0.02,0.16,0.56,1.08,0.15,1.31,0.72,0.15,0.05,1.31,0.14,0.34,1.08,0.02,0,0.1,0.09,0.05,1.36,0.02,0.07,0.53,0.15,0.05,1.25,0.15,0.02,0.1,1.31,0.07,0.56,0.53,1.36,0.15,0.56,0.53,0.34,0.07,0.16,1.31,1.25,0.15 b-chimaerin,8.35,87.38,17.87,189.4,102.8,120.9,87.38,59.44,189.4,52.98,19.93,122.4,87.38,179.2,78.51,30.19,132.2,78.51,62.9,17.87,189.4,132.2,179.2,19.93,0,154.4,149.8,62.9,19.53,19.93,120.9,52.98,78.51,19.75,18.46,78.51,19.93,154.4,17.87,120.9,87.38,102.8,19.53,78.51,19.75,102.8,87.38,120.9,122.4,17.87,18.46,78.51,0.56,1.31,0.14,0.53,0.07,0.56,0.65,0.14,1.69,0.02,0.16,0.56,1.08,0.15,1.31,0.72,0.15,0.05,1.31,0.14,0.34,1.08,0.02,0,0.1,0.09,0.05,1.36,0.02,0.07,1.69,0.15,0.56,1.25,0.15,0.02,0.1,1.31,0.07,0.56,0.53,1.36,0.15,0.56,0.53,0.56,0.07,0.16,1.31,1.25,0.15 unc13,8.52,87.38,17.87,189.4,62.9,59.44,59.44,59.44,189.4,19.53,59.44,122.4,59.44,189.4,78.51,132.2,30.19,78.51,30.19,19.93,154.4,154.4,179.2,19.93,0,154.4,62.9,52.98,19.53,19.93,154.4,52.98,78.51,59.44,19.53,78.51,19.53,120.9,102.8,120.9,87.38,18.46,102.8,78.51,52.98,30.19,154.4,154.4,19.75,62.9,18.46,78.51,0.56,1.31,0.14,0.05,0.65,0.65,0.65,0.14,1.36,0.65,0.16,0.65,0.14,0.15,0.72,1.31,0.15,1.31,0.02,0.1,0.1,1.08,0.02,0,0.1,0.05,1.69,1.36,0.02,0.1,1.69,0.15,0.65,1.36,0.15,1.36,0.07,0.53,0.07,0.56,1.25,0.53,0.15,1.69,1.31,0.1,0.1,0.56,0.05,1.25,0.15 Munc13-1,6.70,87.38,17.87,189.4,30.19,120.9,179.2,59.44,62.9,59.44,59.44,122.4,59.44,132.2,78.51,132.2,30.19,78.51,30.19,19.93,154.4,154.4,179.2,19.93,0,149.8,62.9,52.98,19.53,19.93,132.2,52.98,78.51,19.75,30.19,78.51,19.93,120.9,102.8,78.51,87.38,30.19,102.8,78.51,19.53,18.46,154.4,154.4,17.87,62.9,18.46,78.51,0.56,1.31,0.14,1.31,0.07,1.08,0.65,0.05,0.65,0.65,0.16,0.65,0.72,0.15,0.72,1.31,0.15,1.31,0.02,0.1,0.1,1.08,0.02,0,0.09,0.05,1.69,1.36,0.02,0.34,1.69,0.15,0.56,1.31,0.15,0.02,0.07,0.53,0.15,0.56,1.31,0.53,0.15,1.36,1.25,0.1,0.1,1.31,0.05,1.25,0.15 PKD-C1A,8.60,87.38,62.9,154.4,189.4,120.9,87.38,19.75,132.2,52.98,62.9,122.4,62.9,189.4,78.51,18.46,87.38,78.51,19.93,30.19,132.2,154.4,179.2,19.93,0,154.4,120.9,52.98,19.53,19.93,154.4,102.8,78.51,30.19,19.93,78.51,19.93,154.4,17.87,132.2,87.38,102.8,52.98,78.51,62.9,189.4,102.8,149.8,122.4,17.87,17.87,78.51,0.56,0.05,0.1,0.14,0.07,0.56,0.56,0.72,1.69,0.05,0.16,0.05,0.14,0.15,1.25,0.56,0.15,0.02,1.31,0.34,0.1,1.08,0.02,0,0.1,0.07,1.69,1.36,0.02,0.1,0.53,0.15,1.31,0.02,0.15,0.02,0.1,1.31,0.72,0.56,0.53,1.69,0.15,0.05,0.14,0.53,0.09,0.16,1.31,1.31,0.15 PKD-C1B,8.57,87.38,59.44,189.4,120.9,149.8,87.38,19.75,132.2,59.44,52.98,122.4,59.44,120.9,78.51,19.53,189.4,78.51,102.8,102.8,154.4,154.4,102.8,19.93,0,154.4,189.4,52.98,19.53,19.93,154.4,19.53,78.51,102.8,18.46,78.51,52.98,189.4,17.87,78.51,87.38,102.8,52.98,78.51,62.9,122.4,102.8,120.9,122.4,17.87,17.87,78.51,0.56,0.65,0.14,0.07,0.09,0.56,0.56,0.72,0.65,1.69,0.16,0.65,0.07,0.15,1.36,0.14,0.15,0.53,0.53,0.1,0.1,0.53,0.02,0,0.1,0.14,1.69,1.36,0.02,0.1,1.36,0.15,0.53,1.25,0.15,1.69,0.14,1.31,0.15,0.56,0.53,1.69,0.15,0.05,0.16,0.53,0.07,0.16,1.31,1.31,0.15 rDGKb-C1A,6.71,87.38,120.9,179.2,52.98,154.4,102.8,87.38,189.4,17.87,102.8,122.4,62.9,132.2,78.51,17.87,154.4,78.51,154.4,17.87,132.2,154.4,149.8,19.93,0,120.9,19.93,102.8,19.53,19.93,154.4,78.51,78.51,19.75,189.4,78.51,102.8,132.2,59.44,120.9,87.38,30.19,52.98,78.51,0,62.9,52.98,62.9,122.4,122.4,19.75,78.51,0.56,0.07,1.08,1.69,0.1,0.53,0.56,0.14,1.31,0.53,0.16,0.05,0.72,0.15,1.31,0.1,0.15,0.1,1.31,0.34,0.1,0.09,0.02,0,0.07,0.02,0.53,1.36,0.02,0.1,0.15,0.15,0.56,0.14,0.15,0.53,0.72,0.65,0.07,0.56,1.31,1.69,0.15,0,0.05,1.69,0.05,0.16,0.16,0.56,0.15 hDGKb-C1A,7.87,87.38,120.9,179.2,52.98,154.4,102.8,87.38,189.4,17.87,102.8,122.4,62.9,132.2,78.51,17.87,154.4,78.51,154.4,17.87,132.2,154.4,149.8,19.93,0,120.9,19.93,102.8,19.53,19.93,154.4,78.51,78.51,19.75,189.4,78.51,102.8,132.2,59.44,120.9,87.38,30.19,52.98,78.51,120.9,62.9,52.98,62.9,122.4,122.4,19.75,78.51,0.56,0.07,1.08,1.69,0.1,0.53,0.56,0.14,1.31,0.53,0.16,0.05,0.72,0.15,1.31,0.1,0.15,0.1,1.31,0.34,0.1,0.09,0.02,0,0.07,0.02,0.53,1.36,0.02,0.1,0.15,0.15,0.56,0.14,0.15,0.53,0.72,0.65,0.07,0.56,1.31,1.69,0.15,0.07,0.05,1.69,0.05,0.16,0.16,0.56,0.15 DGKg-C1A,8.55,87.38,62.9,179.2,59.44,132.2,102.8,87.38,189.4,102.8,102.8,122.4,59.44,132.2,78.51,17.87,189.4,78.51,87.38,149.8,132.2,154.4,132.2,19.93,0,120.9,52.98,102.8,19.53,19.93,154.4,78.51,78.51,59.44,132.2,78.51,102.8,132.2,59.44,120.9,87.38,30.19,52.98,78.51,120.9,19.75,52.98,17.87,149.8,122.4,19.93,78.51,0.56,0.05,1.08,0.65,0.34,0.53,0.56,0.14,0.53,0.53,0.16,0.65,0.72,0.15,1.31,0.14,0.15,0.56,0.09,0.34,0.1,0.34,0.02,0,0.07,1.69,0.53,1.36,0.02,0.1,0.15,0.15,0.65,0.72,0.15,0.53,0.72,0.65,0.07,0.56,1.31,1.69,0.15,0.07,0.56,1.69,1.31,0.09,0.16,0.02,0.15
データのPLS回帰分析
偏最小二乗(PLS)回帰分析を使用します.R-GUIを起動し,作業ディレクトリをCSVファイルがある場所に移動します.
[R.app GUI 1.68 (7288) x86_64-apple-darwin13.4.0]
> install.packages("pls")
> library(pls)
> c1 <- read.csv("c1.csv",row.names=1)
分散が0(つまり保存されているアミノ酸残基)の列(column)を削除します(HIS1, PRO11, CYS14, CYS17, GLY23, GLN27, GLY28, CYS31, CYS34, HIS39, CYS42, CYS50; 番号はPKCδ-C1Bの番号)(ひと目でわかる行列(Row ・ Column)の方向の覚え方).
> c1.train <- c1[,apply(c1,2,var) != 0]
PLS回帰分析を行います.pKdが目的変数です.
> c1.pls <- plsr(pKd ~ ., 15, data=c1.train, scale=TRUE, validation="CV")
pKd ~ の後のドット(.)は残り全部という意味.ncompはPLSの潜在変数の最大の数です.scale=Tはオートスケールで,処理の前にデータを平均0,分散1に変換します.CVは交差検証(cross validation)の略です.結果をsummaryで見てみます.
> summary(c1.pls)
Data: X dimension: 27 78
Y dimension: 27 1
Fit method: kernelpls
Number of components considered: 15
VALIDATION: RMSEP
Cross-validated using 10 random segments.
(Intercept) 1 comps 2 comps 3 comps 4 comps 5 comps 6 comps 7 comps 8 comps 9 comps 10 comps 11 comps 12 comps 13 comps 14 comps 15 comps
CV 0.9157 1.185 1.262 1.301 1.168 1.128 1.0501 1.015 0.9908 0.9676 0.9911 1.0024 1.0124 1.0168 1.0115 1.0055
adjCV 0.9157 1.133 1.198 1.233 1.109 1.071 0.9985 0.965 0.9415 0.9179 0.9397 0.9498 0.9587 0.9625 0.9573 0.9516
TRAINING: % variance explained
1 comps 2 comps 3 comps 4 comps 5 comps 6 comps 7 comps 8 comps 9 comps 10 comps 11 comps 12 comps 13 comps 14 comps 15 comps
X 9.171 19.37 28.38 38.39 49.80 55.93 60.77 66.33 69.19 72.99 76.14 78.82 81.81 84.68 87.79
pKd 71.941 86.81 90.92 92.93 94.27 95.62 96.82 97.66 98.68 99.02 99.40 99.73 99.86 99.89 99.91
RMSEPは「予測の平均平方根誤差」(root mean squared error of prediction)の略です.この値が小さい程,予測値とモデルとの乖離が小さいことを表わしています.今回は9つの潜在変数を使用した場合にうまくモデルが構築できているようです.
使用したデータセットのモデルによる予測値と実測値をプロットしてみます(下図).
> plot(c1.train[,1], c1.pls$fitted.value[,,9], xlab="measured pKd", ylab="predicted pKd")
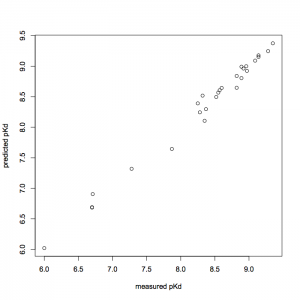
モデルによる予測
MRCK
Serine/threonine-protein kinase MRCKα/β/γの3種のアイソザイムに含まれるC1ドメインの[3H]PDBu結合能を前節で作った回帰モデルで予測してみます(MRCKはmyotonic dystrophy kinase related Cdc42-binding kinaseの略).それぞれの配列は以下の通りです.
MRCKa HQFFVKSFTT PTKCHQCTSL MVGLIRQGCS CEVCGFSCHI TCVNKAPTTC MRCKb HQFSIKSFSS PTQCSHCTSL MVGLIRQGYA CEVCSFACHV SCKDGAPQVC MRCKg HTLRPRSFPS PTKCLRCTSL MLGLGRQGLG CDACGYFCHT TCAPQAPP-C
これらの配列からISAとECIの値を含むCSVファイル(mrck.csv)を作ります.
peptide,pKd,ISA1,ISA2,ISA3,ISA4,ISA5,ISA6,ISA7,ISA8,ISA9,ISA10,ISA11,ISA12,ISA13,ISA14,ISA15,ISA16,ISA17,ISA18,ISA19,ISA20,ISA21,ISA22,ISA23,ISA24,ISA25,ISA26,ISA27,ISA28,ISA29,ISA30,ISA31,ISA32,ISA33,ISA34,ISA35,ISA36,ISA37,ISA38,ISA39,ISA40,ISA41,ISA42,ISA43,ISA44,ISA45,ISA46,ISA47,ISA48,ISA49,ISA50,ISA51,ECI1,ECI2,ECI3,ECI4,ECI5,ECI6,ECI7,ECI8,ECI9,ECI10,ECI11,ECI12,ECI13,ECI14,ECI15,ECI16,ECI17,ECI18,ECI19,ECI20,ECI21,ECI22,ECI23,ECI24,ECI25,ECI26,ECI27,ECI28,ECI29,ECI30,ECI31,ECI32,ECI33,ECI34,ECI35,ECI36,ECI37,ECI38,ECI39,ECI40,ECI41,ECI42,ECI43,ECI44,ECI45,ECI46,ECI47,ECI48,ECI49,ECI50,ECI51 MRCKa,0,87.38,19.53,189.4,189.4,120.9,102.8,19.75,189.4,59.44,59.44,122.4,59.44,102.8,78.51,87.38,19.53,78.51,59.44,19.75,154.4,132.2,120.9,19.93,0,154.4,149.8,52.98,19.53,19.93,78.51,19.75,78.51,30.19,120.9,78.51,19.93,189.4,19.75,78.51,87.38,149.8,59.44,78.51,120.9,17.87,102.8,62.9,122.4,59.44,59.44,78.51,0.56,1.36,0.14,0.14,0.07,0.53,0.56,0.14,0.65,0.65,0.16,0.65,0.53,0.15,0.56,1.36,0.15,0.65,0.56,0.1,0.34,0.07,0.02,0,0.1,0.09,1.69,1.36,0.02,0.15,0.56,0.15,1.31,0.07,0.15,0.02,0.14,0.56,0.15,0.56,0.09,0.65,0.15,0.07,1.31,0.53,0.05,0.16,0.65,0.65,0.15 MRCKb,0,87.38,19.53,189.4,19.75,149.8,102.8,19.75,189.4,19.75,19.75,122.4,59.44,19.53,78.51,19.75,87.38,78.51,59.44,19.75,154.4,132.2,120.9,19.93,0,154.4,149.8,52.98,19.53,19.93,132.2,62.9,78.51,30.19,120.9,78.51,19.75,189.4,62.9,78.51,87.38,120.9,19.75,78.51,102.8,18.46,19.93,62.9,122.4,19.53,120.9,78.51,0.56,1.36,0.14,0.56,0.09,0.53,0.56,0.14,0.56,0.56,0.16,0.65,1.36,0.15,0.56,0.56,0.15,0.65,0.56,0.1,0.34,0.07,0.02,0,0.1,0.09,1.69,1.36,0.02,0.72,0.05,0.15,1.31,0.07,0.15,0.56,0.14,0.05,0.15,0.56,0.07,0.56,0.15,0.53,1.25,0.02,0.05,0.16,1.36,0.07,0.15 MRCKg,0,87.38,59.44,154.4,52.98,122.4,52.98,19.75,189.4,122.4,19.75,122.4,59.44,102.8,78.51,154.4,52.98,78.51,59.44,19.75,154.4,132.2,154.4,19.93,0,154.4,19.93,52.98,19.53,19.93,154.4,19.93,78.51,18.46,62.9,78.51,19.93,132.2,189.4,78.51,87.38,59.44,59.44,78.51,62.9,122.4,19.53,62.9,122.4,0,122.4,78.51,0.56,0.65,0.1,1.69,0.16,1.69,0.56,0.14,0.16,0.56,0.16,0.65,0.53,0.15,0.1,1.69,0.15,0.65,0.56,0.1,0.34,0.1,0.02,0,0.1,0.02,1.69,1.36,0.02,0.1,0.02,0.15,1.25,0.05,0.15,0.02,0.72,0.14,0.15,0.56,0.65,0.65,0.15,0.05,0.16,1.36,0.05,0.16,0,0.16,0.15
Rでこのファイルを読み込みます.
> mrck <- read.csv("mrck.csv", row.names=1)
モデル作成時に削除した行をこちらでも削除します.
> mrck <- mrck[,c(-2,-12,-15,-18,-24,-29,-30,-33,-36,-41,-44,-52,-53,-63,-66,-69,-75,-80,-81,-84,-87,-92,-95,-103)]
モデルで予測します(潜在変数の数は9).
> predict(c1.pls, ncomp=9, newdata=mrck)
, , 9 comps
pKd
MRCKa 7.565233
MRCKb 7.209988
MRCKg 5.848274
MRCKα/β/γの [3H]PDBuに対する予測Kdはそれぞれ,27, 62, 1400 nMでした.MRCKα/βの実測Kdはそれぞれ10.3, 17 nMと報告されています(PKCδ-C1BのKdのそれぞれ58,96倍)(Choi, 2008).今回使用したデータ中のPKCδ-C1BのKdは0.53 nMなので,予測値はそれぞれ51,117倍となり,実験値と傾向がよく一致しました.MRCKγの予測Kd値はμMオーダーなので,MRCKγはphorbol esterに対してinsensitiveであると予想されます.
PKCδ変異体
天然の配列のみから学習したモデルで人為的に変異を入れたドメインの親和性を予測できるのか.Pu(2014) では,PKCδ-C1Bドメインのwild-type, N7R, S10R, P11R, L20RのKdが報告されている(それぞれ,0.23, 1.18, 0.32, 1.19, 8.0 nM).今回のモデルではPRO11が保存されていない配列は予測できないため,それ以外の3種類について予測した.
> predict(c1.pls, ncomp=9, newdata=mrck)
, , 9 comps
pKd
d-C1B_N7R 9.337275 #8.57(補正済実測値)
d-C1B_S10R 9.233776 #9.13
d-C1B_L20R 8.748742 #7.74
あまり一致していなかった.これらの変異体をトレーニングセットに入れて学習させたモデルで予測すると,
pKd d-C1B_N7R 8.943013 #8.57(補正済実測値) d-C1B_S10R 9.084596 #9.13 d-C1B_L20R 7.602420 #7.74
と(当然ですが)先程より一致しました.トレーニングセットにどのデータを使うかで,予測精度は大きく変わります.
HumanとRatの配列の違い
PKCδ-C1A,δ-C1B,およびη-C1Bはヒトとラットで配列にわずかに違いがあります.
PKCd-C1A(rat) HEFIATFFGQPTFCSVCKEFVWGLNKQGYKCRQCNAAIHKKCIDKIIGRC PKCd-C1A(human) HEFIATFFGQPTFCSVCKDFVWGLNKQGYKCRQCNAAIHKKCIDKIIGRC PKCd-C1B(rat) HRFKVYNYMSPTFCDHCGTLLWGLVKQGLKCEDCGMNVHHKCREKVANLC PKCd-C1B(human) HRFKVHNYMSPTFCDHCGSLLWGLVKQGLKCEDCGMNVHHKCREKVANLC PKCh-C1B(rat) HKFNVHNYKVPTFCDHCGSLLWGIMRQGLQCKICKMNVHIRCQANVAPNC PKCh-C1B(human) HKFSIHNYKVPTFCDHCGSLLWGIMRQGLQCKICKMNVHIRCQANVAPNC
この違いがリガンド親和性に影響するかどうか予測.
> round(predict(c1.pls,ncomp=9,newdata=human),2)
, , 9 comps
pKd
d-C1A_human 7.31
d-C1B_human 9.22
h-C1B_human 9.28
d-C1A_rat 7.32
d-C1B_rat 9.25
h-C1B_rat 9.37
References
- Irie, K.; Nakahara, A.; Ohigashi, H.; Fukuda, H.; Wender, P. A.; Konishi, H.; Kikkawa, U. Synthesis and phorbol ester-binding studies of the individual cysteine-rich motifs of protein Kinase D. Bioorg. Med. Chem. Lett. 1999, 9, 2487–2490. DOI: 10.1016/S0960-894X(99)00413-8. [PMID]: 10498194.
- Shindo, M.; Irie, K.; Masuda, A.; Ohigashi, H.; Shirai, Y.; Miyasaka, K.; Saito, N. Synthesis and phorbol ester binding of the cysteine-rich domains of diacylglycerol kinase (DGK) isozymes. J. Biol. Chem. 2003, 278, 18448–18454. DOI: 10.1074/jbc.M300400200. [PMID]: 12621060.
- Irie, K.; Masuda, A.; Shindo, M.; Nakagawa, Y.; Ohigashi, H. Tumor promoter binding of the protein kinase C C1 homology domain peptides of RasGRPs, chimaerins, and Unc13s. Bioorg. Med. Chem. 2004, 12, 4575–4583. DOI: 10.1016/j.bmc.2004.07.008. [PMID]: 15358285.
- Qian, Y.; Liang, Y.; Liu, W.; Liang, G. Comprehensive comparison of twenty structural characterization scales applied as QSAM of antimicrobial dodecapeptides derived from Bac2A against P. aeruginosa. J. Mol. Graph. Model. 2017, 71, 88–95. DOI: 10.1016/j.jmgm.2016.11.003. [PMID]: 27863328.
- Collantes, E. R.; Dunn, W. J. 3rd. Amino acid side chain descriptors for quantitative structure-activity relationship studies of peptide analogues. J. Med. Chem. 1995, 38, 2705–2713. DOI: 10.1021/jm00014a022. [PMID]: 7629809.
- Choi, S. H.; Czifra, G.; Kedei, N.; Lewin, N. E.; Lazar, J.; Pu, Y.; Marquez, V. E.; Blumberg, P. M. Characterization of the interaction of phorbol esters with the C1 domain of MRCK (myotonic dystrophy kinase-related Cdc42 binding kinase) α/β. J. Biol. Chem. 2008, 283, 10543–10549. DOI: 10.1074/jbc.M707463200. [PMID]: 18263588.
- Pu, Y.; Kang, J. H.; Sigano, D. M.; Peach, M. L.; Lewin, N. E.; Marquez, V. E.; Blumberg, P. M. Diacylglycerol lactones targeting the structural features that distinguish the atypical C1 domains of protein kinase C ζ and ι from typical C1 domains. J. Med. Chem. 2014, 57, 3835–3844. DOI: 10.1021/jm500165n. [PMID]: 24684293.
(了)